- Home
- About
- Departments
- Research Activities
Advanced Research Center
Established in 2020
- Research Support Units
Co-ordinates All Activities Of The Institute
- Resources
Diarrhoea Surveillance
Lead Investigator(s)
No data was found
Diarrhoea Surveillance

Lead Investigator
Prof. Dorothy Yeboah-Manu
Project Summary
Surveillance and Laboratory Support for Emerging Diarrhoea Pathogens
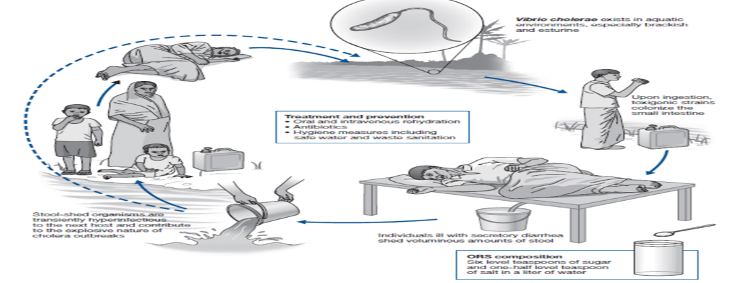
Diarrheal disease remains a major global health problem particularly in children under five years and the emergence of antibiotic-resistant strains of causative pathogens could slow control efforts particularly in settings where treatment options are limited. AS a department, we have conducted two major studies on bacterial derived diarrheal. The first study aimed at investigating Vibrio cholerae isolates from cholera outbreaks in southern Ghana using molecular epidemiology. Cholera remains a major global public health threat and continuous emergence of new V. cholerae strains is of major concern. Due to the annual cycles of cholera in Ghana, NMIMR in collaboration with National Public health Reference Laboratory conducted a molecular epidemiological study to detect virulence markers and antimicrobial resistance patterns of V. cholerae isolates obtained from the 2012–2015 cholera outbreaks in Ghana. The study sought to phenotypically characterize archived enterobacterial isolates from suspected cholera cases, determine the antibiotic sensitivity pattern of isolated V. cholerae using Kirby Bauer disc diffusion, identified major genomic virulent markers by PCR and strain clonality by multi-locus variable number tandem repeat analysis (MLVA). We identified some antibiotics that are no longer effective in the treatment of the disease, new circulating strains (Inaba serotype and non-O1/O137 serogroups) and some diarrhea-causing bacteria other than V. cholerae that also play a role in the annual cholera outbreaks. We also found that the V. cholerae strains circulating in Ghana harbor several virulent genes and each year, a unique clone predominates the cholera outbreak. This research identified Ablekuma sub-district as the major hot spot for cholera.
Our second study was a multidisciplinary study involving diarrhoea surveillance conducted in Ghana aimed to determine the antimicrobial susceptibility profile of diarrhea-causing bacteria and viruses. For the bacteria arm, our objectives were; 1) To determine various microbial pathogens involved in diarrhoeal cases in Ga-West Municipal, 2) To determine the antibiotic sensitivity pattern and resistant conferring molecular markers to inform appropriate case management, and 3) To genotype pathogens identified for assessment of risk factors and prediction of possible outbreaks. We observed significant findings including the detection of several different bacteria etiology of diarrhoea aside V. cholerae which include, Salmonella, Shigella, Vibrio sp (parahaemolyicius, albensis), and Enterotoxigenic E. coli (ETEC). We also observed high resistance of isolates to commonly abused drugs such as tetracycline and noticed that multi-drug resistant Vibrio sp and extended spectrum beta-lactamase producing E. coli have the capability of destabilizing our health systems if drugs administered are not effective.
Project Background
Surveillance and Laboratory Support for Emerging Diarrhoea Pathogens
Diarrheal disease remains a major global health problem particularly in children under five years and the emergence of antibiotic-resistant strains of causative pathogens could slow control efforts particularly in settings where treatment options are limited. AS a department, we have conducted two major studies on bacterial derived diarrheal. The first study aimed at investigating Vibrio cholerae isolates from cholera outbreaks in southern Ghana using molecular epidemiology. Cholera remains a major global public health threat and continuous emergence of new V. cholerae strains is of major concern. Due to the annual cycles of cholera in Ghana, NMIMR in collaboration with National Public health Reference Laboratory conducted a molecular epidemiological study to detect virulence markers and antimicrobial resistance patterns of V. cholerae isolates obtained from the 2012–2015 cholera outbreaks in Ghana. The study sought to phenotypically characterize archived enterobacterial isolates from suspected cholera cases, determine the antibiotic sensitivity pattern of isolated V. cholerae using Kirby Bauer disc diffusion, identified major genomic virulent markers by PCR and strain clonality by multi-locus variable number tandem repeat analysis (MLVA). We identified some antibiotics that are no longer effective in the treatment of the disease, new circulating strains (Inaba serotype and non-O1/O137 serogroups) and some diarrhea-causing bacteria other than V. cholerae that also play a role in the annual cholera outbreaks. We also found that the V. cholerae strains circulating in Ghana harbor several virulent genes and each year, a unique clone predominates the cholera outbreak. This research identified Ablekuma sub-district as the major hot spot for cholera.
Our second study was a multidisciplinary study involving diarrhoea surveillance conducted in Ghana aimed to determine the antimicrobial susceptibility profile of diarrhea-causing bacteria and viruses. For the bacteria arm, our objectives were; 1) To determine various microbial pathogens involved in diarrhoeal cases in Ga-West Municipal, 2) To determine the antibiotic sensitivity pattern and resistant conferring molecular markers to inform appropriate case management, and 3) To genotype pathogens identified for assessment of risk factors and prediction of possible outbreaks. We observed significant findings including the detection of several different bacteria etiology of diarrhoea aside V. cholerae which include, Salmonella, Shigella, Vibrio sp (parahaemolyicius, albensis), and Enterotoxigenic E. coli (ETEC). We also observed high resistance of isolates to commonly abused drugs such as tetracycline and noticed that multi-drug resistant Vibrio sp and extended spectrum beta-lactamase producing E. coli have the capability of destabilizing our health systems if drugs administered are not effective.
Objectives & Research Areas
Key Findings
Ongoing Activities
No data was found



A community durbar was held on the 25th of January 2022 in Joma, a small town under the Ga West Municipal assembly to educate all community stakeholders on the pathogens causing diarrhoea and how to minimize infection rates.
Dr. David Opare
Dr. Samuel Duodu
Prof. William Ampofo
Virology
Dr. Evelyn Bonney
Virology
Francine Ntoumi,
Université Marien NGouabi, Fondation Congolaise pour la Recherche Médicale (FCRM), Brazzaville, Congo, Institute for Tropical Medicine, University of Tübingen, Tübingen, Germany
Alimuddin Zumla,
Division of Infection and Immunity, University College London and NIHR Biomedical Research Centre, UCL Hospitals NHS Foundation Trust, London, England, United Kingdom
Hiroshi Kiyono
The Institute of Medical Science, The University of Tokyo, Tokyo, Japan
Dr. Asiedu Bekoe
Ghana Health Service
The Pan-African Network for Rapid Research, Response, Relief and Preparedness for Infectious Diseases Epidemics (PANDORA-ID-NET) Consortium
AMED-JICA (the Science and Technology Research Partnership for Sustainable Development, SATREPS)